Smart Cancer Detection
Using a Convolutional Neural Network to Search 3D Medical Images
About the Project
The objective of this project was to predict the presence of lung cancer given a 40×40 pixel image snippet extracted from the LUNA2016 medical image database. This problem is unique and exciting since it will have a direct impact on the future of health care, computer vision in medicine, and how machine learning will affect personal decisions. The medical field is a likely place for machine learning to thrive, as regulations continue to allow increased sharing of anonymous data for the sake of better care. It's even more exciting since machine learning in medicine is still new enough that our project is able to implement methods at the forefront of technology!
Skills Involved
- TensorFlow
- TensorBoard
- Convolutional Neural Networks
- Hyperparameter tuning
- Medical image processing
- Python
Procedure
Due to the complex nature of our task, most machine learning algorithms are not well-posed for this project. There are currently two prominent approaches for machine learning for image data: either extract features using conventional computer vision techniques and learn the feature sets, or apply convolution directly using a CNN. In the past few years, however, CNNs have far outpaced traditional computer vision methods for difficult, enigmatic tasks such as cancer detection. We decided to implement a CNN in TensorFlow, Google’s machine learning framework.
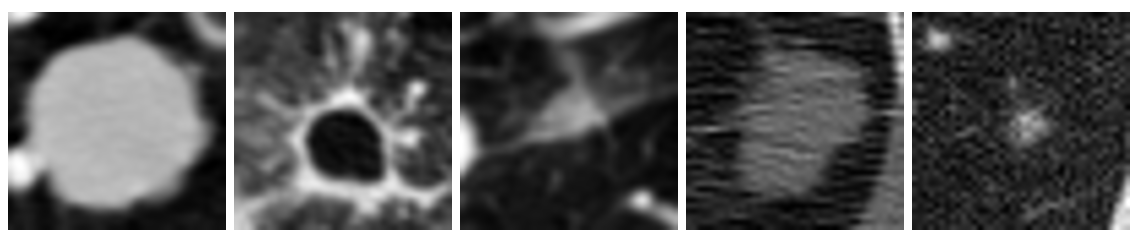
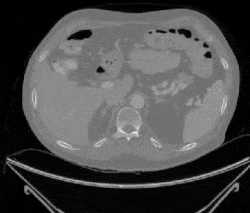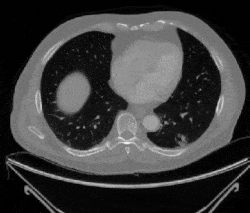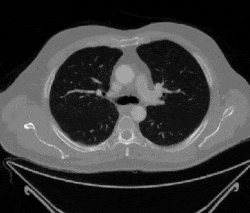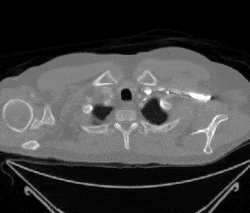
Figure 1: Examples of cancerous images
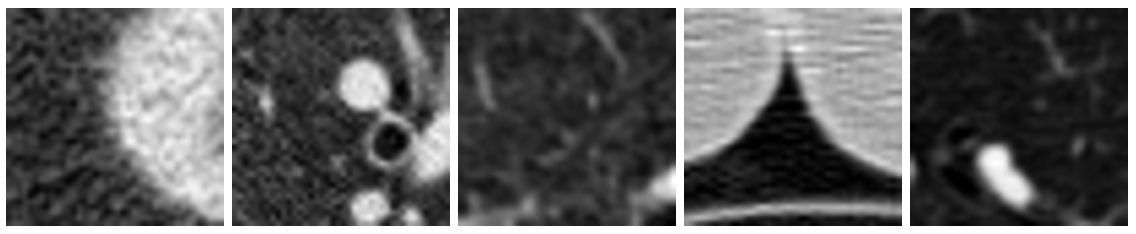
Figure 2: Examples of non-cancerous images
Because my partners and I had limited experience with convolutional neural networks, we elected to first explore our hyperparameter space using a large-scale experiment. We ran tests across 5 of the dimensions for a total of 108 unique models. For this study, we kept a constant network architecture, although it was varied for other runs.
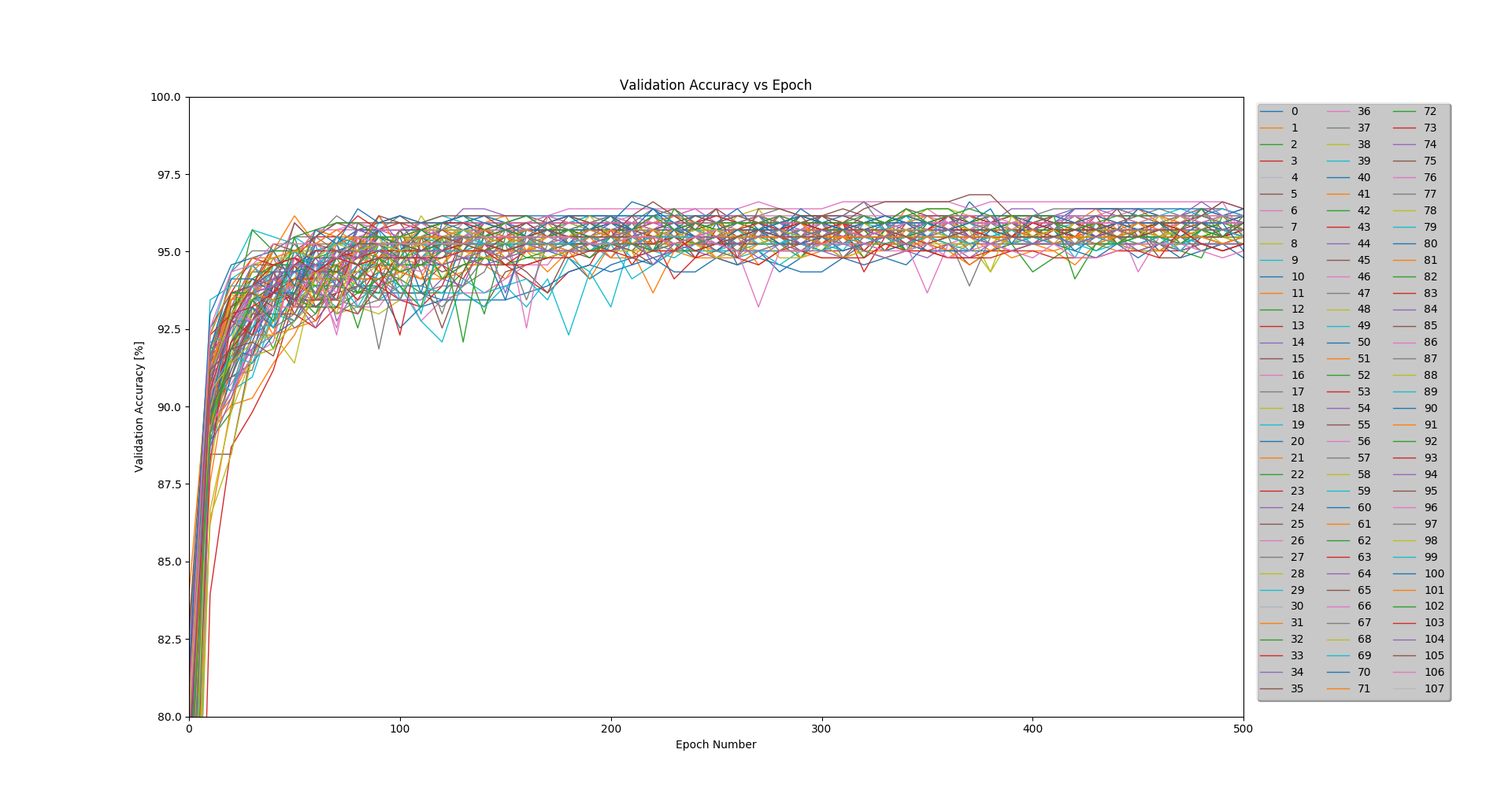
Figure 3: Accuracy results for the 108 runs of the experiment
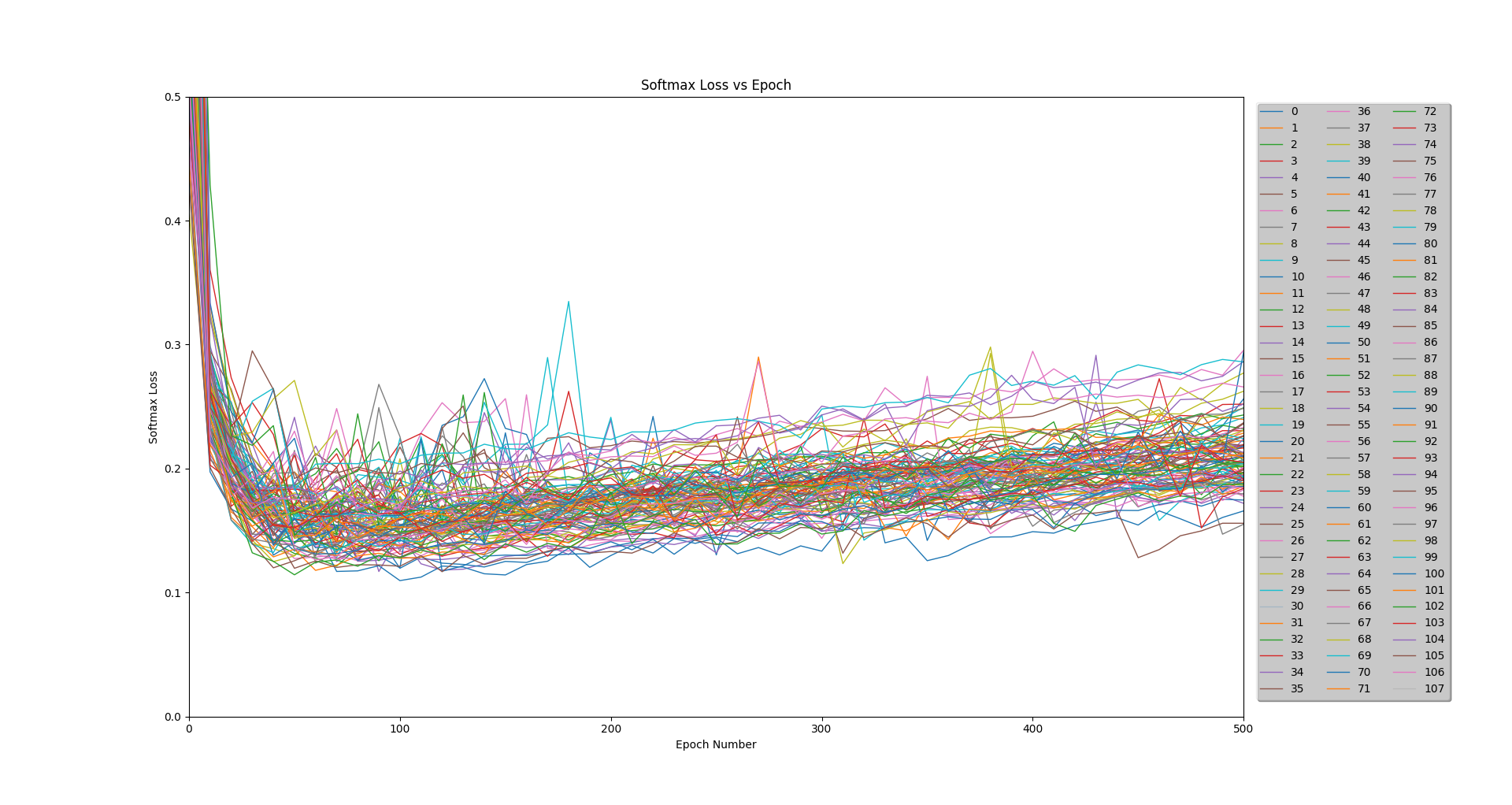
Figure 4: Loss results for the 108 runs of the experiment
Each model was trained on 2,064 images using a batch size of 104. A validation test was run every 10 epochs on a separate 442 images, and a final test was run after 500 epochs on a remaining 442 images.
Table 1: Hyperparameter Permutations
| Attribute | Values Tested |
|---|---|
| Convolutional Layer 1: Kernel Size | 3x3, 5x5, 7x7 |
| Convolutional Layer 1: Number of Filters | 16, 32 |
| Convolutional Layer 2: Kernel Size | 3x3, 5x5, 7x7 | Convolutional Layer 2: Number of Filters | 32, 64 |
| Dropout Rate | 10%, 20%, 30% |
Results
After determining the best set of hyperparameters based on average peak validation accuracy, we then tested six new architectures based on these hyperparameters. The structure of each of these architectures was decided based on the principles described in the Stanford CS231n course notes. After running the final six architectures at 500 epochs, we found the inflection point of the loss to be around 250 epochs. We then ran each of the six architectures for 250 epochs and recorded the final test accuracy. The best network architecture of these six achieved a test accuracy of 96.38%.
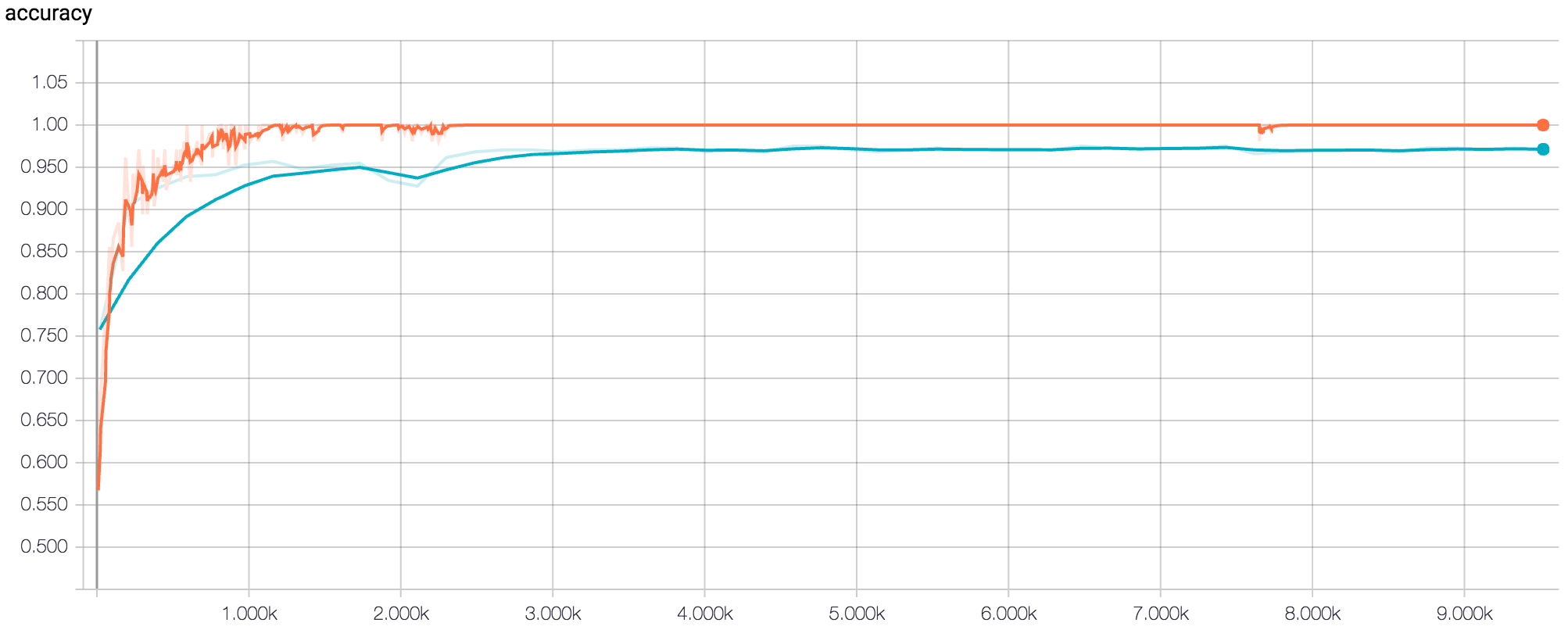
Figure 5: TensorBoard Graph of accuracy for final model after 500 epochs (orange = training, blue = validation)
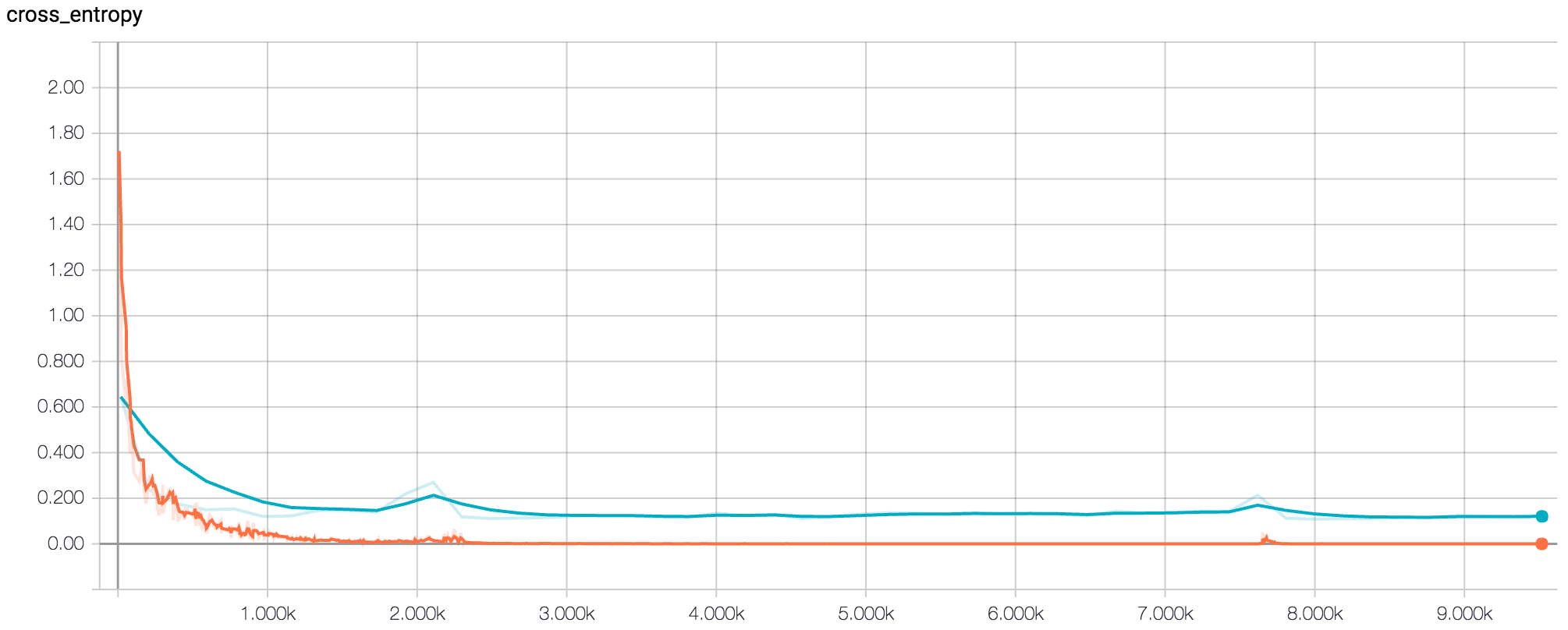
Figure 6: TensorBoard Graph of (softmax) loss for final model after 500 epochs (orange = training, blue = validation)
Conclusions
After choosing the model with the highest accuracy and best robustness, we dove into the results to learn why some images were misclassified (CNNs are capable of nearly 100% classification, even for extremely complex tasks). The first step was to construct a confusion matrix (see table below) to determine if there was a clear trend in misclassification.
Table 2: Confusion Matrix
| Predicted Positive | Predicted Negative | |
|---|---|---|
| Actual Positive | 226 | 12 |
| Actual Negative | 4 | 200 |
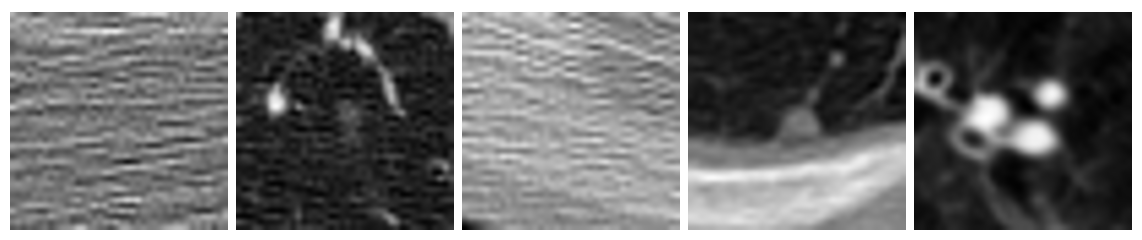
Figure 7: Examples of misclassified images from the test dataset
While the model did produce more false negatives than positives, we believe the problem isn't intrinsic to the model. If you look at the misclassified images above, it's easy to see that some are completely non-descript. Even trained radiologists would have trouble classifying these images without more information. It's possible there were some mis-labeled images in the dataset, or perhaps the tumor was too large to fit in the 40×40 window we chose. Furthermore, we discovered that all of the top 5 models consistently and unanimously misclassified the same images. We believed it would be against the spirit of the exercise to remove these images from the dataset, but are certain we could acheive near 100% accuracy if these problematic images were eliminated.
In the future, we hope to test our model on full 3D lung scans. A tool like this could search through millions of lung scans per day and assist radiologists by pinpointing potential nodules, marking suspicious areas, and clearing patients with no signs of cancerous. It could also run checks on patients with no symptoms, without patients needing to pay for multiple hours of these specialists' time!
Learn More
This project fulfilled my final requirement for EECS 349: Machine Learning, but went well beyond class requirements, diving deep into my and my partners' interests in machine learning for computer vision. If you're interested in learning more, take a look at our final report. The report and all of our source code can be downloaded from GitHub.